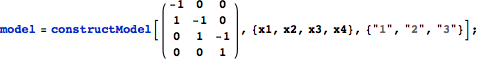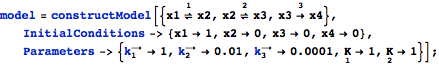Model construction and manipulation
The MASS Toolbox relies on a model structures
Model construction
| constructModel[{rxn1,rxn2, ...}] | construct a model from a list of reactions |
| constructModel[S,{m1,m2,...},{id1,id2,...}] | construct a model from stoichiometry matrix S, metabolites m and reaction identifiers |
Constructing models.
Using a list of reactions to construct a model.
| Out[9]= |  |
Using a stoichiometry matrix construct a model.
Model attributes
Models provide a series of attributes (see
Model attributes for more information about individual attributes). Some of them are mutable, e.g, InitialConditions or Parameters, others are immutable, e.g., Species or Stoichiometry. Immutable attributes usually depend on other mutable attributes and thus can be modified indirectly.
| model["Attributes"] | returns a list of all available attributes in model |
| getAttribute[model] | returns the respective attribute |
| model["Attribute"] | returns the respective attribute |
| setAttribute[model, new] | replaces the respective attribute entirely with new |
| updateAttribute[model, new] | adds non-existing information |
Handling model attributes.
Get a list of all available attributes.
| Out[15]= |  |
Initially, the model does not contain any initial conditions or parameters.
| Out[16]= |  |
| Out[17]= |  |
Using
getter functions to access the same attributes.
| Out[18]= |  |
| Out[19]= |  |
Using
setter functions to include parameters and initial conditions. The model is changed in place.
Initial conditions and parameters have been properly set.
| Out[22]= |  |
| Out[23]= |  |
Attributes can also be provided during the model construction process.
| constructModel[args,Attr1 →{...}, ...] | provide additional information during model construction |
Provide attribute information during model construction.
Set initial conditions and parameters during model construction
| Out[25]= |  |
Using
updater functions to modify attributes
| Out[28]= |  |
Add and delete reactions
| deleteReactions[model, {r1,r2,...}] | return new model with reactions deleted |
| addReactions[model, {r2,r2, ...}] | return new model reactions added |
Change reaction content.
| Out[36]= |  |
Delete the added reaction
| Out[38]= |  |
Set operations
| Union[model, model2, ...] | Unify models |
| Intersection[model, model2, ...] | Get the intersection of models |
| Complement[model, model2, ...] | Get the complement of models |
Set operations for models.
Get the union of two model
| Out[34]= |  |